Text Classification 3 Ways – Logistic Regression, Random Forests, and XGBoost
I've been planning to write this post for a while, but I've debated about a few things. First, many posts about text classification or sentiment analysis use simple binary classifiers and do something like classifying tweets or Yelp reviews as positive or negative. What about classifying into multiple classes? There are some examples out there, but not many. Second, the posts typically use nice, neat datasets that don't require much data/file manipulation. This is not how life is when working with actual production data. Third, I was not sure if I wanted to experiment on finding optimal hyperparameters for each classification algorithm on my dataset or just give examples of how to perform the overall process. I chose the later. If you want a fun project, take these Jupyter Notebooks and the dataset and do grid search or other methods for hyperparameter optimization. Maybe I'll do that in another post.
In this post, I'll discuss how I performed text classification on Pubmed abstracts using logistic regression, random forests, and XGBoost. In each case, I stuck with pretty much default hyperparameters, so the accuracy for each is good, but could definitely be improved.

You may not be familiar with Pubmed. "PubMed® comprises more than 30 million citations for biomedical literature from MEDLINE, life science journals, and online books. Citations may include links to full-text content from PubMed Central and publisher web sites." It is a huge dataset! I decided to use abstracts from these documents as my dataset. I chose 5 categories of articles and downloaded 2,000 abstracts for each. The categories were:
1-Dental caries. These are cavities in teeth.
2-Dental pulp. The inner part of the tooth.
3-Endodontic treatment. Root canals are included in this.
4-Periodontal disease. Disease of the gums.
5-Transposable elements. Repeated sequences of DNA in chromosomes. I studied these for my PhD dissertation (it's a fascinating read) so they are near and dear to me.
I chose these categories for 2 reasons. First, they were similar (at least the first 4) and I wanted to see how well the classifiers would work on similar categories with many overlapping terms. Second, transposable elements was a complete outgroup, so it should be able to classify them easily.
The process I used for classification is very straightforward. You can check out the Jupyter Notebooks to see the actual code.
1-Download the abstracts from Pubmed. They were in what is called "Pubmed" format and had to be parsed. It wasn't a nice clean dataset.
2- Read the parsed datasets and show some basic summary stats.
3-Preprocess each abstract - remove section headings and other unneeded characters, lowercase, tokenize, lemmatize, and stem all text. Describing all of this is beyond this post, but there are many good resources to describe them.
4-Create TF-IDF word vectors. Other options could be used, but I chose to use this one because it is straightforward.
5-Split train/test data.
6-Create the classifier and train the model.
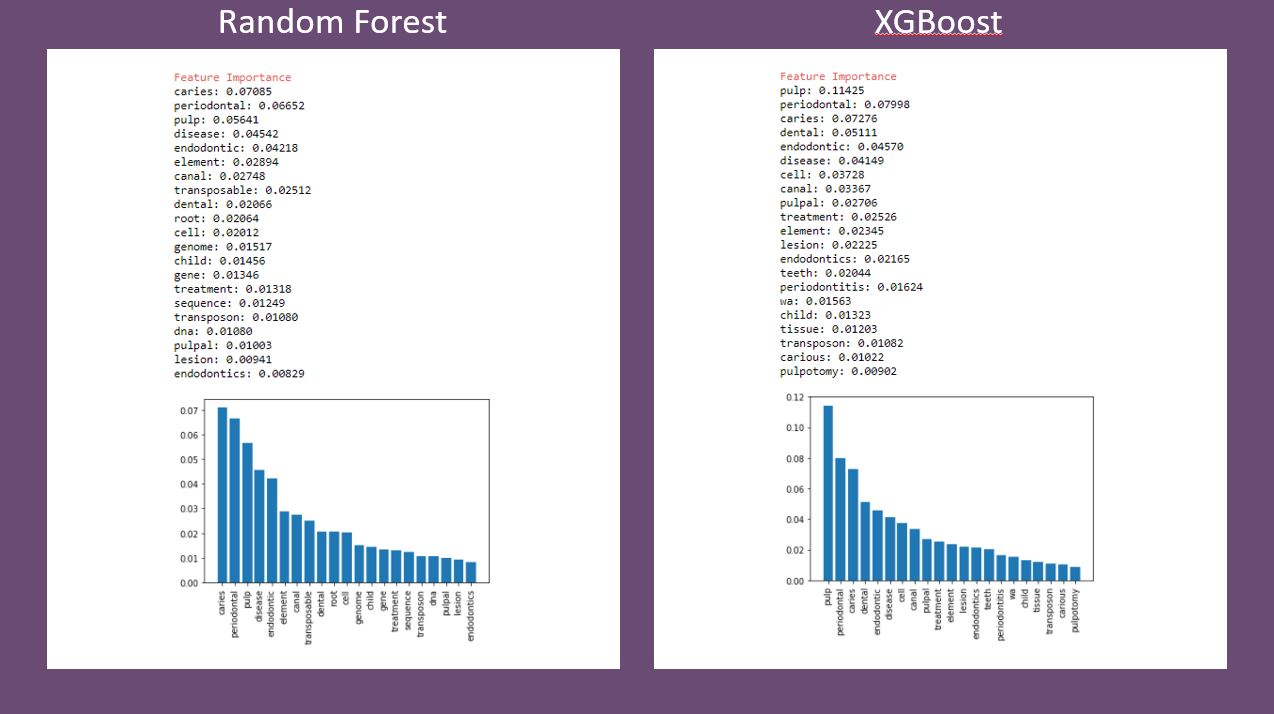
7-For random forest and XGBoost, find the most important features in the model and display them.
8-Print the metrics - accuracy, classification report, and confusion matrix.
9-Classify previously unseen abstracts using the newly created model.
The notebooks, along with the data files, are found on my Github. Each notebook is the same for steps 1-5, but differs in the actual classifier created and trained. Even with default hyperparameters, the results were not bad and were consistent across all classifiers. Below is a comparison of feature importance as determined by random forest and XGBoost. There are some similarities and some differences. Again, this would change, and most likely be even more similar, with hyperparameter optimization.
Here are the accuracy values on the test data:
- Random forest - 96.048%
- Logistic regression - 96.698%
- XGBoost - 95.398%
Classification of the unseen abstracts was good as well. For a simple quick and dirty analysis, this is the way to go. Test each out, then experiment with the hyperparameters. Training time for each classifier is different, with XGBoost taking by far the longest. Take this into consideration. If you have a nice server to run it on, that will definitely help! I hope you will take a few minutes and check out the Notebooks.
