MONAI User & Onboarding Guide (AI Toolkit for Medical Imaging)
MONAI Core offers dataset handling features on top of PyTorch making training models and segmentation tasks using medical data easier. Examples include functions to help import common medical file types and transformation functions that allow for easy dataset augmentation.
Below are the steps required to run some of MONAI’s provided tutorial scripts which are located on this page.
MONAI Core Installation:
The Jupyter Notebook we will be running is provided by MONAI and uses the MedNIST dataset to train a model to perform image classification. The model’s goal is to determine if the given image is of a Hand, an Abdomen CT, a CXR, a Chest CT, a Breast MRI, or a HeadCT.
First we need to start by installing MONAI on our system. Then we will be able to check that the installation was successful by running our first sample Jupyter Notebook. To install MONAI and all other packages you need for this notebook, run the following pip install commands:
pip install "MONAI-weekly[gdown, nibabel, tqdm, itk]"
pip install sklearn matplotlib
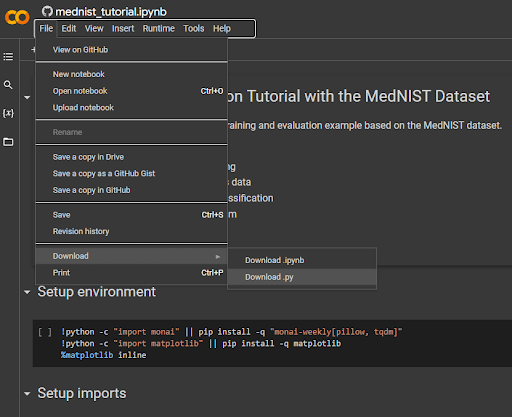
Transfer the downloaded file (should be mednist_tutorial.py)to the machine on which you installed MONAI if it’s not there already. Comment out or delete the three lines shown in the screenshot above in the Setup environment section of the script. Then run the script using Python (ie python3 mednist_tutorial.py)
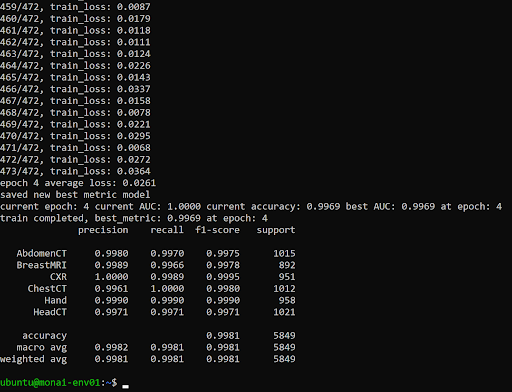
This should kick off the training of an image classification model. This will take several minutes to complete. The output when it’s finished running successfully should look like the screenshot below.
Note: If you get a “CUDA out of memory” error, change the batch size on lines 169, 172, and 175 to a smaller number such as 100
MONAI Core 3D Segmentation Tutorial Script:
The process for running a 3D segmentation script is much the same. This time we will be using the spleen segmentation notebook MONAI provides. The link to the notebook hosted on Google Colab is here. First download the .py script from the Colab version of this notebook just like with the previous one and then comment out or delete the three lines in the Setup environment section of the notebook. Then, transfer the script to the machine and run it using Python (ie python3 spleen_segmentation_3d.py) .
You should be able to run the other tutorial scripts MONAI provides in following this process. Since MONAI provides a good variety of scripts, this is a good starting point for exploring the capabilities of MONAI Core.
